Auckland outbreak: the genomes
Marc Daalder at newsroom has a very good piece about genome sequencing and its role in handling the Auckland outbreak.
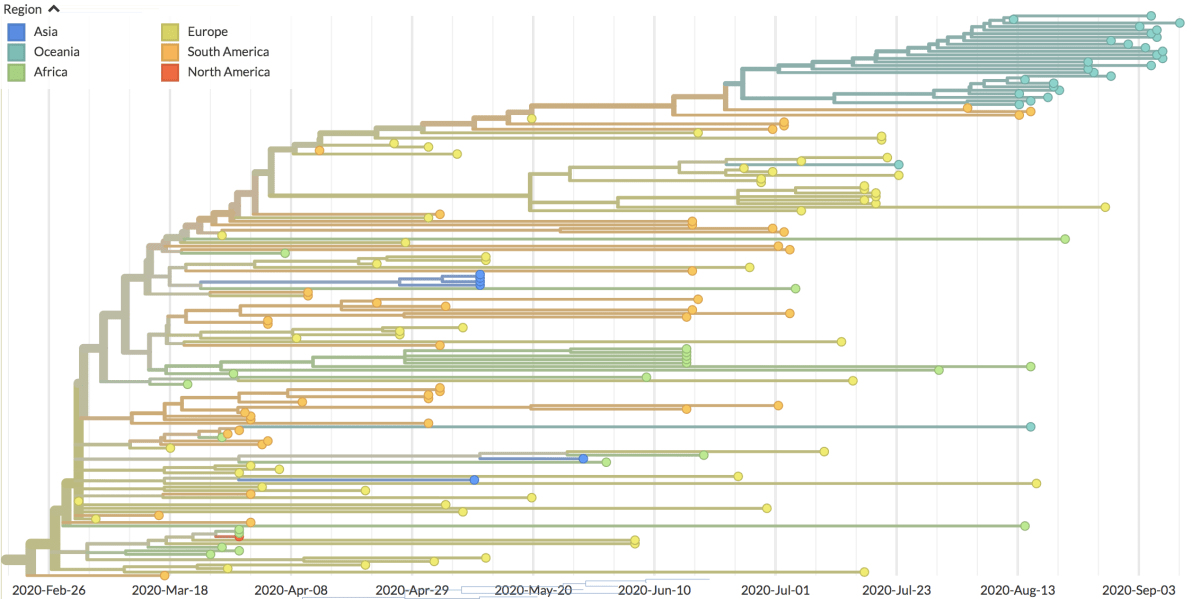
One thing I want to highlight is this family tree. It’s the B.1.1.1 clade of the virus, a particular viral subfamily
The samples from the Auckland outbreak are that blue/green cluster at the top. They’re all more closely related to each other than to any other sample that has been sequenced in NZ or anywhere else in the world, so we know they had a recent common ancestor: the virus that started the outbreak.
I’m mentioning this because I saw discussion on Twitter over the past month or so of the ‘B.1.1.1 clade’ and whether there had been other earlier cases from that clade in NZ and whether that meant the government was hiding something. B.1.1.1 is a pretty broad family, going back to a common ancestor in late February. If two samples are from different broad clades, they are different incursions to NZ, but if they’re from the same broad clade they aren’t necessarily the same incursion. You need the full sequence to say how closely viruses are related.
Thomas Lumley (@tslumley) is Professor of Biostatistics at the University of Auckland. His research interests include semiparametric models, survey sampling, statistical computing, foundations of statistics, and whatever methodological problems his medical collaborators come up with. He also blogs at Biased and Inefficient See all posts by Thomas Lumley »